RLA plot
Loading...
PCA plot
Loading...
SD plot
Loading...
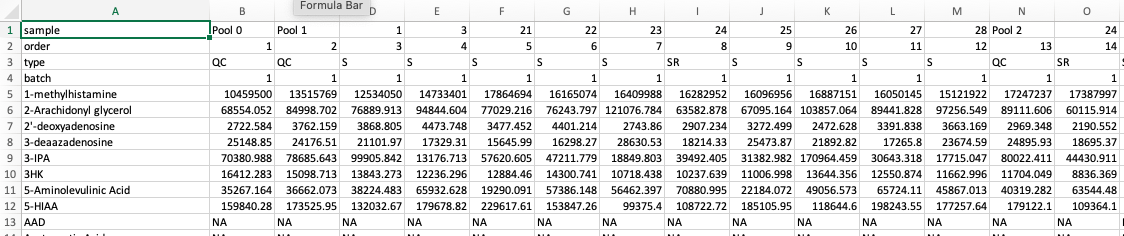
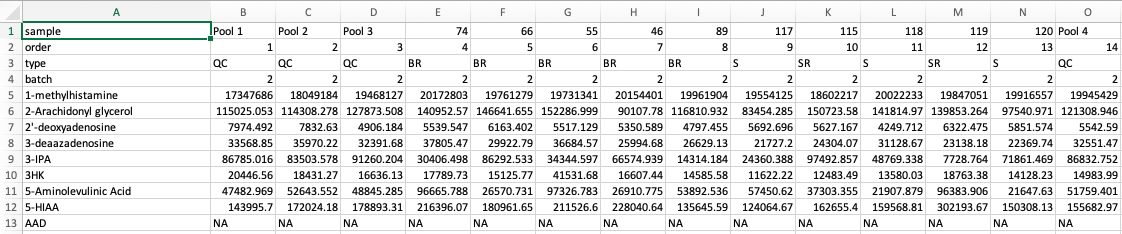
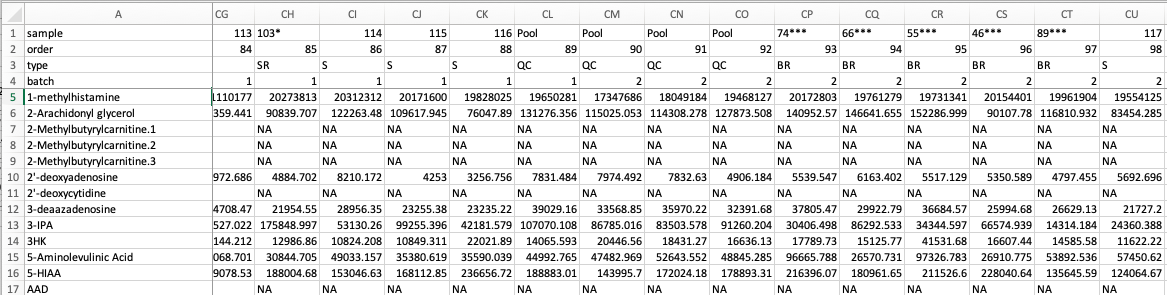
hRUV shiny app expects multiple Comma Separated Values (CSV) files. There are two possible formats to submit to hRUV. Select the option from the side bar to see an example format.
Note: Make sure the first 4 rows of the first column (cell A1-4) match the names as shown in the image below. (I.e. In specific order of 'sample', 'order', 'type' and 'batch'
Each CSV file should only contain samples from the same batch. The file structure should be in the format as below:
^ : Equal contribution
^ : Equal contribution